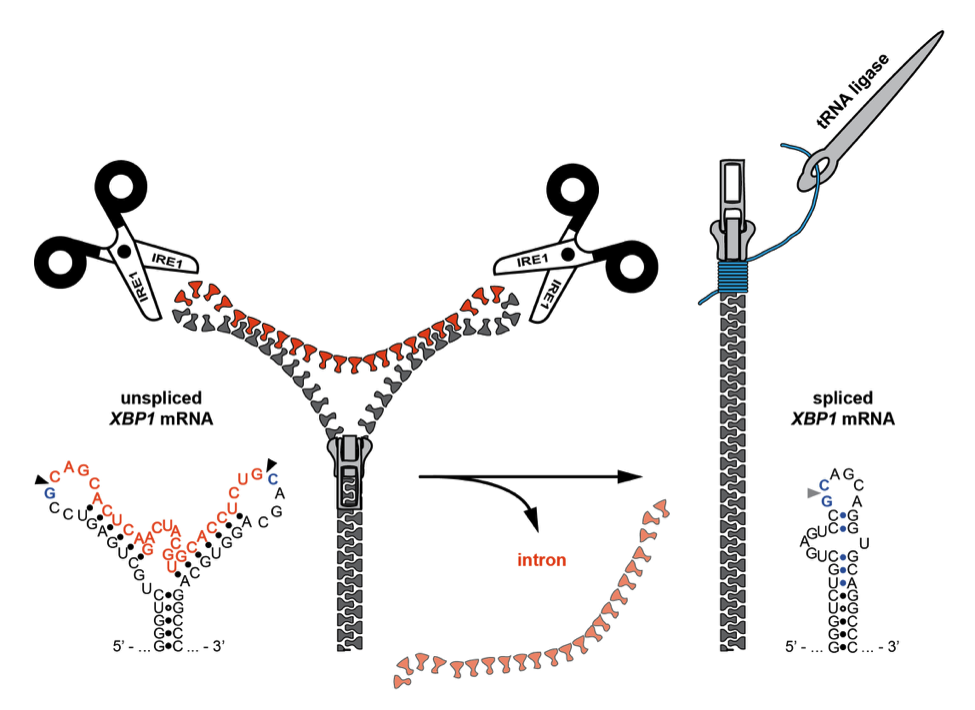
Ire1-mediated signaling during the unfolded protein response encompasses fascinating aspects of basic RNA biology including mRNA targeting, cleavage, and turnover as well as mRNA quality control.
Protein folding stress in the ER activates the cytosolic endonuclease domain of the transmembrane stress sensor Ire1. The activated endonuclease initiates a stress response through two distinct modes of enzymatic action: i) by catalyzing the first step in a non-conventional mRNA splicing reaction and ii) by degrading mRNAs at the ER surface through regulated Ire1-dependent decay (RIDD). For splicing, the substrate mRNA is actively targeted to the Ire1 signaling sites, and then cleaved at the splice sites. Next, a unique RNA conformational change actively ejects the intron and coordinates the two exons, which are efficiently ligated by the RNA processing enzyme tRNA ligase. This unique cytosolic splicing reaction is, unlike conventional splicing in the nucleus, entirely catalyzed by protein enzymes. We aim to understand the precise orchestration of the mRNA processing steps at the ER membrane during the UPR.
While mRNA splicing leads to the production of a transcription activator that induces UPR target genes to enhance the protein folding capacity of the ER, in RIDD Ire1 cleaves and destabilizes a set of ER-localized mRNAs and thus reduces the protein folding burden entering the ER. RIDD is initiated by the activation of Ire1 and requires additional factors involved in ribosome rescue as well as exonucleases that degrade the fragmented mRNA. Both functional outputs depend on the sequence- and structure-dependent cleavage of ER-localized mRNAs by Ire1. In metazoans, both mRNA processing mechanism, RIDD and splicing, are present. We are interested in understanding the regulation of both functional outputs and their distinct roles in regulating ER homeostasis. Interestingly, the fission yeast S. pombe exclusively uses RIDD, whereas the budding yeast S. cerevisiae exclusively uses Ire1-dependent mRNA splicing. We are cross-exchanging the machinery and RNA substrates between these organisms to learn where the mechanistic differences lie.