As subcellular compartments, organelles provide optimized conditions for distinct biochemical reactions. Explained by their endosymbiotic origin, mitochondria and chloroplasts retain their own unique genomes. However, the organellar genomes have limited coding capacities compared to the nuclear genomes, which encodes the majority of organellar proteins. Therefore, proper biogenesis and function of mitochondria and chloroplasts requires crosstalk between spatially separated compartments. As such, organelle inheritance and integrity is faithfully and continuously monitored.
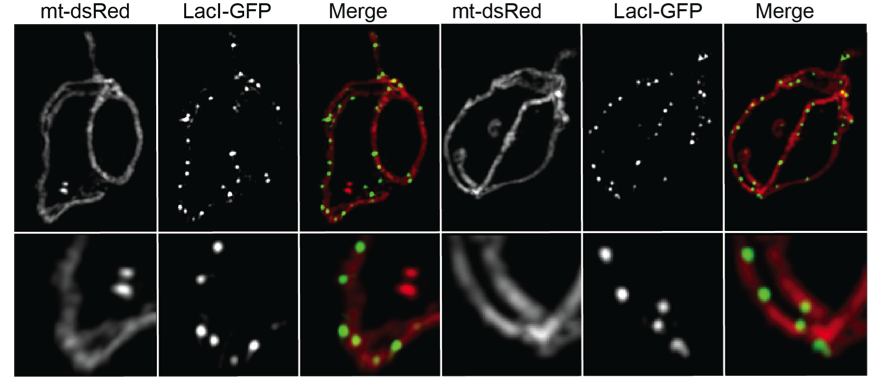
In our lab we maintain a long-standing interest in the mechanisms that determine the morphology and dynamics of mitochondria and the distribution and inheritance of the mitochondrial genome (mtDNA). Some 20 years ago, we showed that mitochondria in the yeast, S. cerevisiae, form a single interconnected network that is maintained by balanced fusion and fission events. This observation paved the way for the identification of the machinery involved in these processes. A fascinating advance came with the discovery of the ERMES (ER-mitochondria encounter structure) complex that rivets mitochondria to the endoplasmic reticulum at sites of fission and actively replicating mitochondrial genomes. To gain mechanistic insights into the role of the ER in mtDNA inheritance, we developed a system to fluorescently label and follow mitochondrial genomes in living cells by video microscopy. Such observations lead to the discovery that mtDNA is segregated to the mitochondrial tips generated by mitochondrial fission and offer insights into the mechanisms of mtDNA segregation and distribution. Remarkably, mtDNA copy number is tightly controlled in cells and regulated in response to different metabolic conditions. We developed a genetic screen in S. cerevisiae to identify mutants that alter mtDNA levels. With this screen we identified a yet uncharacterized protein as a candidate regulator of mtDNA copy number and are in the midst of deciphering how it functions.
Tail-anchored (TA) proteins are a family of membrane proteins that have a single hydrophobic sequence at their extreme C-terminus tail. Their targeting and insertion into the ER membrane uses dedicated molecular machinery, comprising the GET pathway (TRC40 in mammals). Some GET-targeted proteins integrate into mitochondrial membranes inappropriately, necessitating their removal. Recently, our lab identified Msp1 (ATAD1 in mammals), a conserved, membrane-anchored AAA ATPase that senses mistargeted TA proteins to mitochondria and extracts them for degradation. When Msp1 is disrupted, functional and morphological defects ensue, underscoring that Msp1 plays a crucial role in achieving fidelity of OMM TA protein targeting. We are exploring the molecular mechanism by which Msp1 accomplishes this feat.
Recently, a chloroplast unfolded protein response (cpUPR) was discovered in the green algae Chlamydomonas reinhardtii that is also conserved in higher plants. This pathway allows photosynthetic eukaryotes to sense the presence of damaged proteins in chloroplasts and to mitigate the resulting stress by reprogramming nuclear gene expression. Conceptually, the cpUPR is similar to UPRs operating from the endoplasmic reticulum and the mitochondria. However unlike for these responses, the components that transmit the cpUPR signal from chloroplasts to the nuclear gene expression system remain to be identified. We conducted a forward genetic screen in C. reinhardtii and isolated a collection of mutants that are defective in signaling. Excitingly, we mapped the mutation of the strongest silencing cpUPR mutants to a single gene, MARS1, that encodes an uncharacterized serine/threonine kinase. This result marks the identification of the first molecular component of the cpUPR and opens the door to its mechanistic dissection and the identification of up- and downstream players in the pathway.