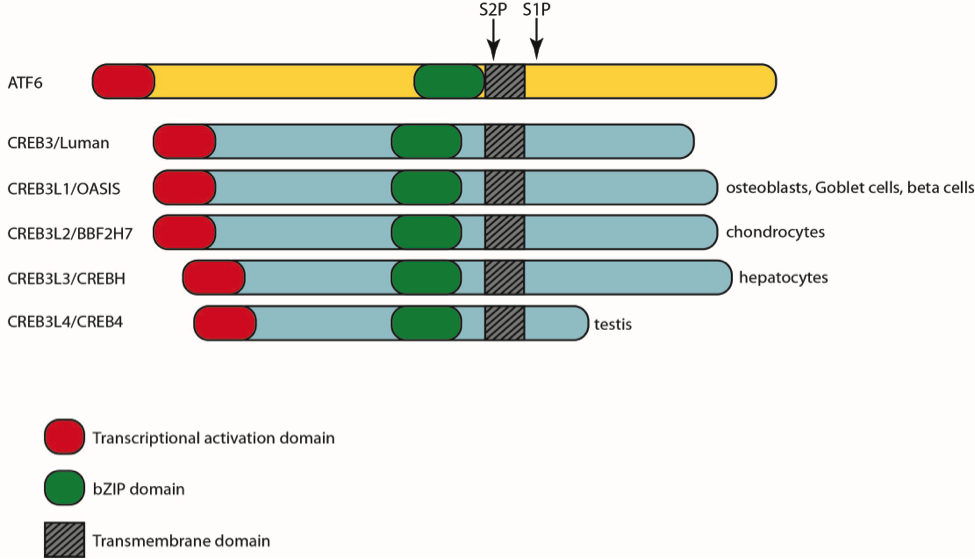
One family of proteins that senses conditions in the ER and increases the organelle’s folding capacity is comprised of a family of membrane-bound transcription factors, named after the founding member ATF6 (activating transcription factor). These proteins have a stress-sensing domain localized in the ER, a transmembrane domain, and a cytosolic transactivating domain (Figure 1). The ATF6 family has seven members, the ATF6 paralogs, ATF6α and ATF6β, and five CREB3 proteins – CREB3, CREB3L1, CREB3L2, CREB3L3, CREB3L4. ATF6α and ATF6β are ubiquitously expressed and respond to all ER stress; loss of both ATF6α and ATF6β is incompatible with life. The CREB3 proteins show tissue specific expression in secretory tissues and are thought to play more specialized roles. For example, CREB3L1 is primarily expressed in bone cells. Loss of CREB3L1 is a cause of osteogensis imperfecta, a disease associated with defective collagen, the primary component of the secreted bone matrix.
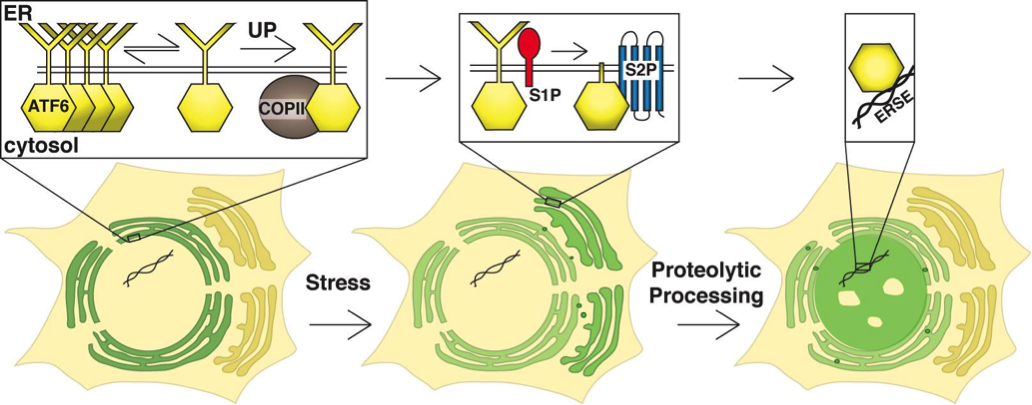
To control gene expression, these transcription factors must first be released from the membrane. The mechanism by which these proteins sense stress remains a mystery and we are actively pursuing these questions in our lab. Activation of ATF6 and family members requires regulated intramembrane proteolysis, where their cytosolic transactivating domains are freed from their membrane anchors. In unstressed cells, ATF6 is sequestered in the ER (Figure 2). Upon sensing ER stress, ATF6 family members are released from the ER and traffic to the Golgi apparatus where they encounter two proteases, site-1 and site-2 proteases respectively. The released cytosolic domain then translocates to the nucleus, binds its transcriptional co-activators and induces expression of its target genes. For ATF6α these are chaperones and lipid synthesis genes that increase both the size and functional capacity of the ER. The target genes of CREB3 proteins are not well established but appear to vary depending on the cell type in which they are expressed. We do not understand how ATF6 family members are sequestered in the ER in unstressed cells or how they are triggered to rapidly move to the Golgi apparatus upon ER stress.
To investigate how ATF6 responds to stress, we study ATF6α. We generated a series of cell-based assays that monitor both the subcellular localization and the transcriptional output of ATF6α. We used these assays to screen a library of small molecules and found one scaffold that blocks activation of ATF6α by ER stress. We called this scaffold Ceapin, from the Irish verb “ceap” meaning “to trap”, as in the presence of Ceapin ATF6α remains trapped in the ER and cannot signal. Ceapin selectively inhibits ATF6α but not the other family members that use the same trafficking machinery and proteases for activation, including its close paralog ATF6β. We currently aim to identify the target protein to which Ceapin binds to exert its effects and use Ceapin to interrogate the genetic circuitry and protein interactome of ATF6α to find the missing pieces of the activation machinery. We are extending our studies to other ATF6 family members.