The unfolded protein response maintains protein homeostasis in the endoplasmic reticulum (ER), where the majority of transmembrane and secretory proteins are modified, folded and assembled. When the ER-resident folding machinery becomes overwhelmed, misfolded polypeptides accumulate and lead to ER stress, with toxic consequences to cells. To adjust the protein-folding capacity of the ER according to the needs of the cell, ER-resident sensor/transducer proteins constantly monitor the protein folding status in the ER lumen and take corrective actions to maintain proper ER function. This collection of conserved signaling pathways is termed the unfolded protein response (UPR).
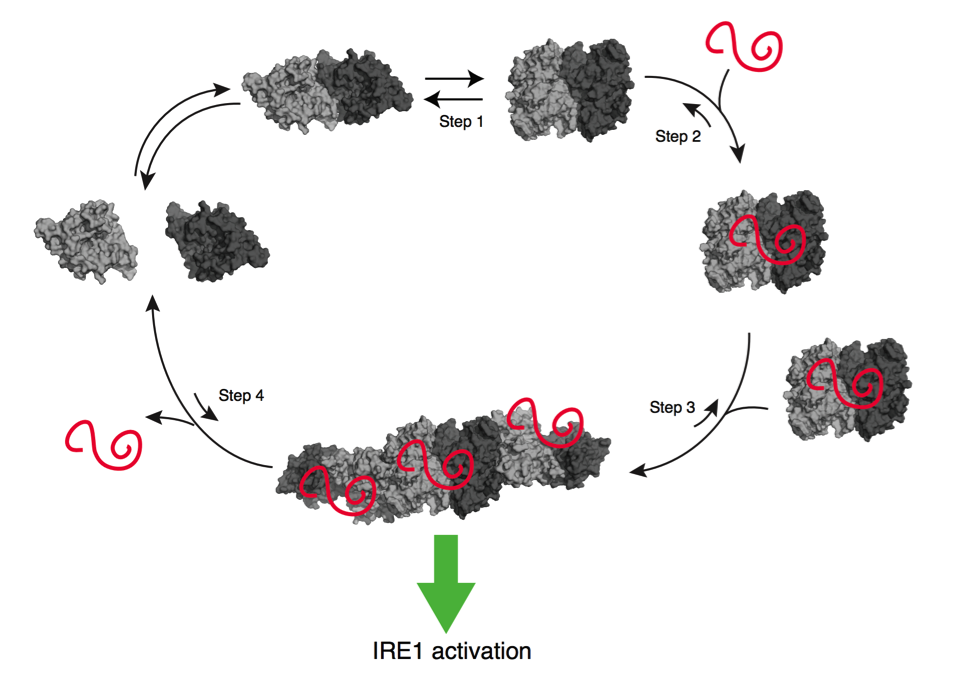
Since its discovery some 20 years ago, our knowledge of the UPR has vastly expanded. Many of the initial insights were obtained from the budding yeast Saccharomyces cerevisiae, where a single signal transduction pathway is tasked with responding to ER stress. This pathway is initiated by the stress sensor/transducer IRE1, a highly conserved kinase/endoribonuclease that lies within the ER membrane and responds to the accumulation of unfolded proteins in the ER lumen. In higher eukaryotes, IRE1’s endonuclease activity results in two distinct outcomes: a transcriptional response program initiated by the non-conventional splicing of the mRNA encoding the potent transcription factor XBP1, and the general decrease in protein load through degradation of diverse mRNAs (regulated IRE1-dependent decay, or RIDD). Our lab is interested in the structural and spatial determinants of IRE1’s selectivity towards its many mRNA targets, as well as the general principles behind post-transcriptional regulation via mRNA processing. IRE1 is also known to form large homo-oligomers in a stress-dependent way. The functional significance of these oligomers, as well as their spatial context, is a subject of ongoing investigation in the lab. Finally, we lack a comprehensive structural understanding of the recognition of unfolded proteins by IRE1’s lumenal domain. We seek to address these questions through a combination of structural, biochemical, and imaging-based approaches.
Defects in UPR are also linked to a number of cancers and protein misfolding disorders and can potentially be therapeutically targeted. For example, upon unmitigated ER stress, IRE1 activity attenuates and shifts the UPR towards a pro-apoptotic outcome. Understanding how the cell makes this life-or-death decision under stress is crucial for pharmacologic modulation of the UPR in disease conditions. In a recent collaboration with the Ashkenazi lab at Genentech, we found that ER stress-induced apoptosis acts through Death Receptor 5 (DR5), whose mRNA transcript is a substrate of RIDD. The deactivation of RIDD allows for the upregulation of DR5, which induces apoptosis through a novel intracellular mechanism. We are currently investigating how ER stress triggers intracellular DR5 oligomerization and activation.